Our work on Automated Gleason Grading in Lancet Oncology
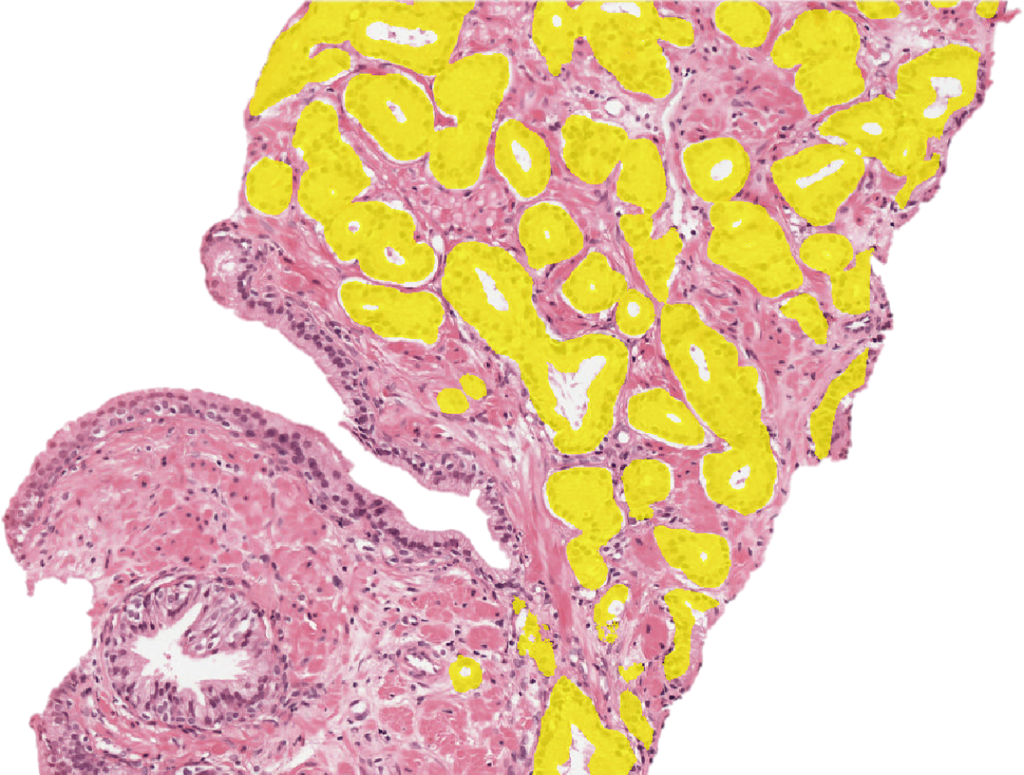
With 1.2 million new prostate cancer cases each year, a high incidence-to-mortality ratio, and risk of overdiagnosis and overtreatment, there is a strong need for accurate assessment of patient prognosis. Currently, the Gleason score, assigned by a pathologist after microscopic examination of cancer morphology, is the most powerful prognostic marker for prostate cancer patients. However, it suffers from significant inter- and intra-observer variability, limiting its usefulness for individual patients.
As an alternative, Wouter Bulten et al developed a fully-automated cancer detection and Gleason grading system for entire prostate biopsies. The system was developed using 5759 biopsies from 1243 patients. A semi-automatic labeling technique was used to circumvent the need for manual pixel-level annotations. The study was focussed on the full range of Gleason grades, and evaluated on a large cohort of patients with an expert consensus reference standard and an external tissue microarray test set. Their work was accepted for publication by The Lancet Oncology and appeared online today.
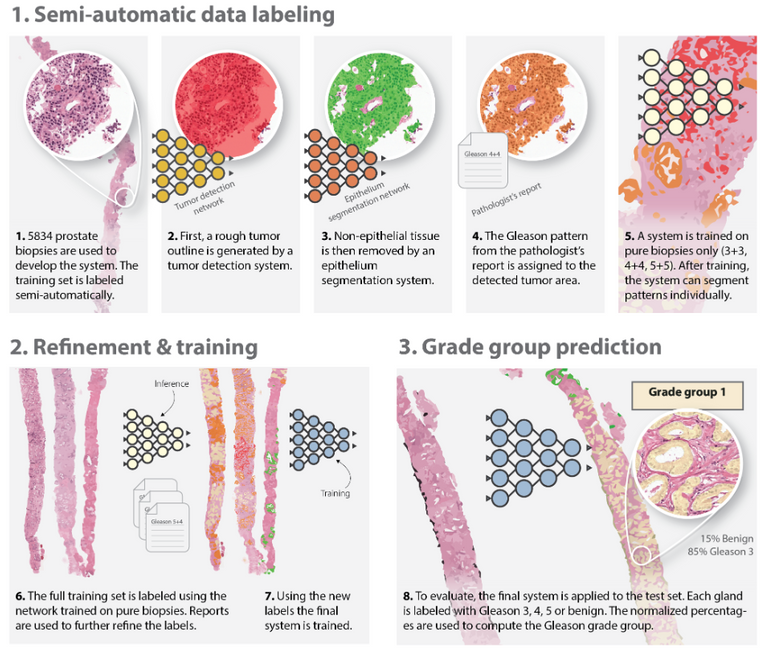
The figure above depicts the development of the deep learning system. The authors employed a semi-automated method of labelling the training data (top row), removing the need for manual annotations by pathologists. The final system can assign Gleason growth patterns on a cell-level.
The developed system achieved a high agreement with the reference standard (quadratic kappa 0.918). In a separate observer experiment, the deep learning system outperformed 10 out of 15 pathologists in agreement with the reference standard. The system was validated on an external test set where it achieved an AUC of 0.977 on distinguishing between benign and malignant biopsies and an AUC of 0.871 using grade group 2 as a cut-off.
Click here to try Wouter's algorithm on your own data and learn more about the project on automated Gleason grading.
← Back to overview