Streaming CNNs for Multi-Megapixel Images
Graphics Processing Units (GPUs) are fundamental for training deep Convolutional Neural Networks (CNNs). They typically have about 11GB in memory, and up to 24GB for higher end GPUs. However, this is not enough to train the CNNs on full resolution digital pathology images to identify the presence of disease accurately. Hans Pinckaers et al. from the Diagnostic Image Analysis Group have now found a solution to this problem and their work was accepted for publication by IEEE Transactions on Pattern Analysis and Machine Intelligence.
They developed a novel method to directly train state-of-the-art convolutional neural networks using any input image size in an end-to-end manner. This method exploits the locality of most operations in modern CNNs by performing the forward and backward pass on smaller tiles of the image. In this work, they showed a proof of concept using images of up to 66-megapixels (8192x8192), saving approximately 50GB of memory per image.
Memory demand is typically highest in the first few layers of state-of-the-art CNNs before several pooling layers are applied because the intermediate activation maps are large. These activation maps require much less memory in subsequent layers. They proposed to construct these later activations by streaming the input image through the CNN in a tiled fashion, changing the memory requirement of the CNN to be based on the size of the tile and not the input image. This method allows the processing of input images of any size.
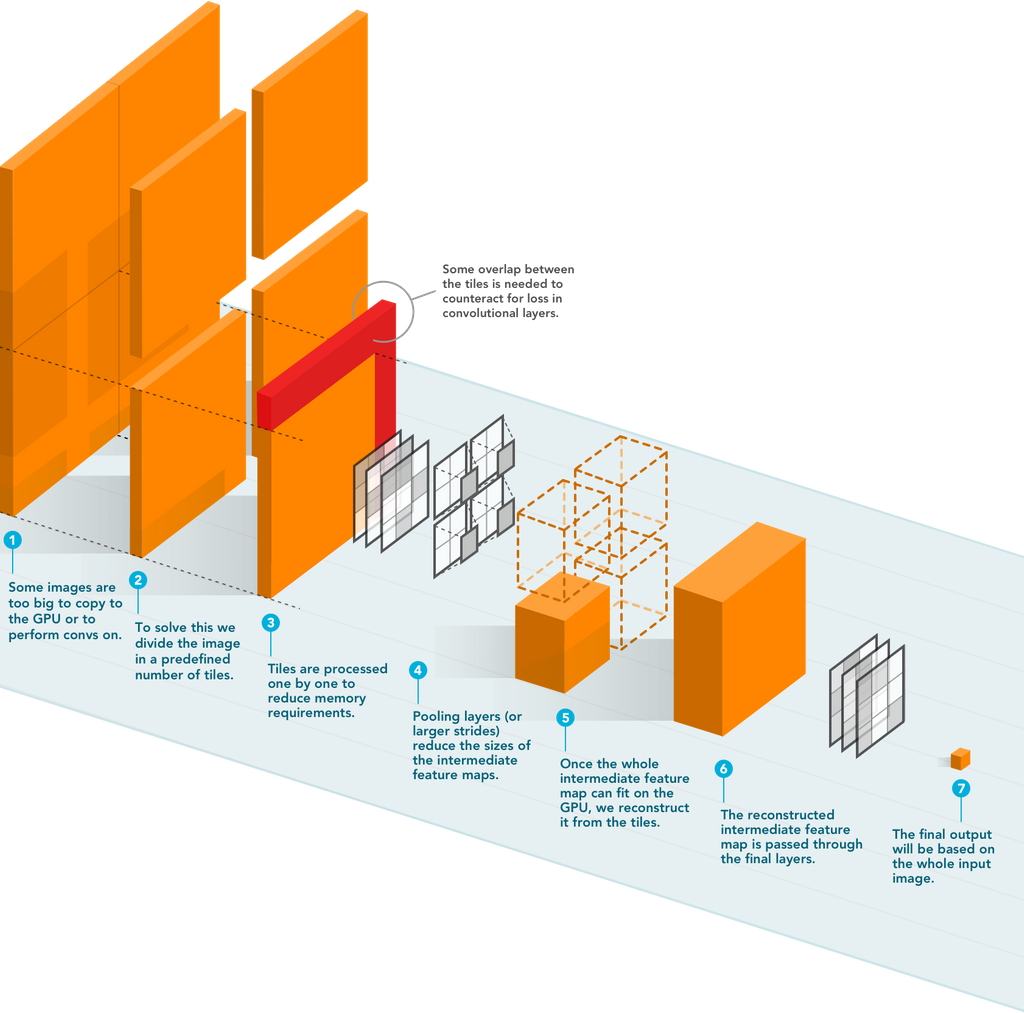
The figure above depicts the schematic overview of streaming. A detailed explanation of the algorithm can be found in their paper.
They improved the AUC from 0.580 on 4-megapixel images to 0.706 on 66-megapixel images for metastasis detection in breast cancer on the CAMELYON17 dataset.
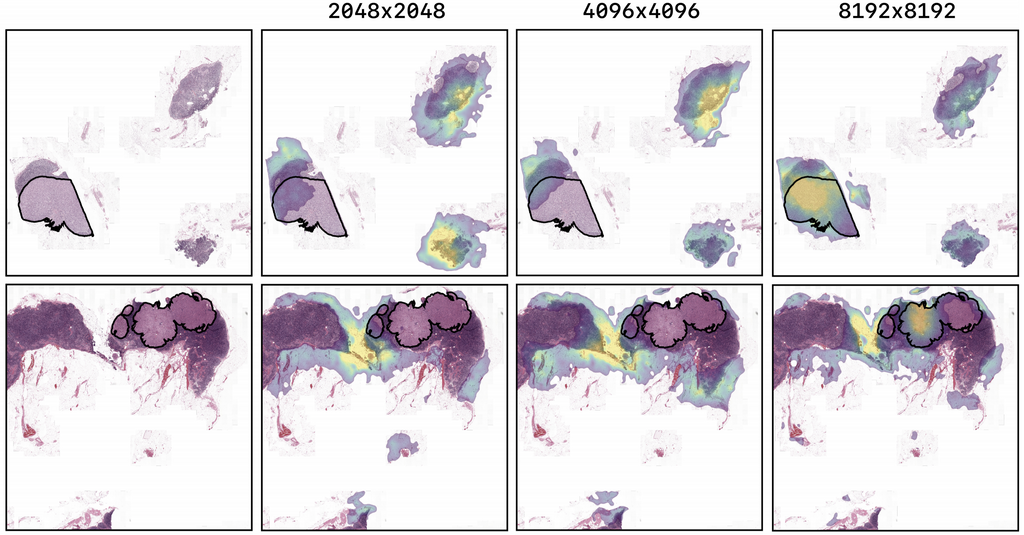
The figure above shows the saliency maps for images of the tuning set of the CAMELYON17 experiment. The highest resolution model, trained on image-level labels shows highlights corresponding to the ground truth pixel-level annotation of a breast cancer metastasis. The lower resolution models have lower probability for the ground truth class and show little correspondence to the location of the metastases.
They also obtained a Spearman correlation metric approaching state-of-the-art performance on the TUPAC16 dataset, from 0.485 on 1-megapixel images to 0.570 on 16-megapixel images.
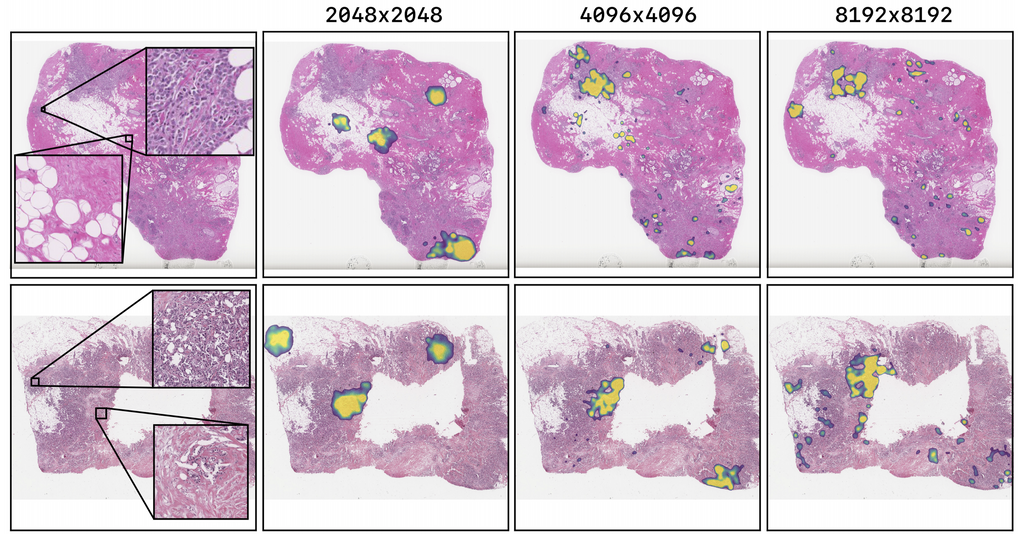
The figure above shows the saliency maps for test set images of the TUPAC16 experiment using the best performing models. The TUPAC16 network shows highlights in cell-dense and cancerous regions. There is a trend in which the higher the input solution of the model, the less it focuses on healthy tissue. Also, higher resolution models focus on more locations of the tissue.
The code to reproduce the experiments is publicly available here.
← Back to overview