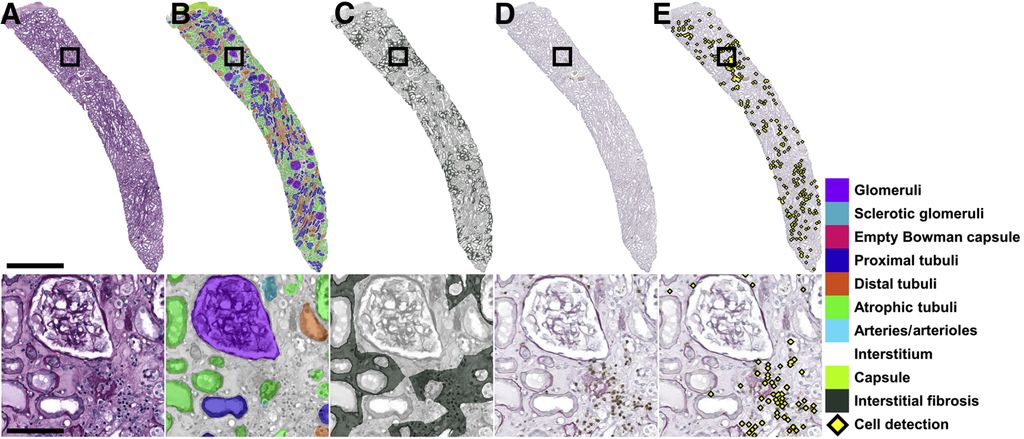
Clinical Problem
Inflammation and chronic changes in the different tissue compartments (e.g. glomeruli, tubulo-interstitium, vasculature) are important prognostic features for kidney (transplant) function and guide treatment decision. Quantitative scoring of these features is therefore very important for transplant diagnostics and kidney research. However, visual scoring by pathologists is poorly reproducible and labor-intensive. Within the DIAGGRAFT project (funded by the Dutch kidney foundation), we aim to automatically quantify inflammation in different tissue compartments. This is only possible if we have a reliable tissue segmentation.
Approach
Hermsen et al.[1] have proposed a first approach to the task of tissue segmentation using a U-net architecture. This approach, however, did not include peritubular capillaries (PTC), for which we have since collected annotations. The goal of this project is to extend the tissue segmentation model to include this additional class and to investigate current state-of-the-art segmentation model(s) to improve the performance.
Data
The dataset used in[1] contains 125 whole slide images (WSI) annotated for 10 different tissue types (glomeruli, sclerotic glomeruli, empty Bowman capsules, proximal tubuli, distal tubuli, atrophic tubuli, capsule, arteries/arterioles, interstitium, and border). Additionally, there is a dataset of 69 WSI with annotations for peritubular capillaries. If needed, more images are available, and a resident pathologist is involved in the project to help interpret results and provide new annotations.
References
[1] Hermsen, Meyke, et al. "Convolutional neural networks for the evaluation of chronic and inflammatory lesions in kidney transplant biopsies." The American Journal of Pathology 192.10 (2022): 1418-1432.
[2] Jayapandian, Catherine P., et al. "Development and evaluation of deep learning–based segmentation of histologic structures in the kidney cortex with multiple histologic stains." Kidney international 99.1 (2021): 86-101.
[3] Zhang, Jingwei, et al. "Sam-path: A segment anything model for semantic segmentation in digital pathology." International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer Nature Switzerland, 2023.
[4] Liu, Shilong, et al. "Grounding dino: Marrying dino with grounded pre-training for open-set object detection." arXiv preprint arXiv:2303.05499 (2023).
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Interest in image analysis and machine learning.
- Affinity with programming in Python, specifically in the PyTorch framework.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- You will be part of the Diagnostic Image Analysis Group (DIAG) and Computational Pathology Group, whose research focus is the analysis of histopathological slides with deep learning techniques.
- You will have access to and work with a large GPU cluster.
- For more information, please contact Linda Studer


