Background
There is a specific type of cancer occurring throughout the gastrointestinal (GI) tract called signet ring cell carcinoma (SRCC), based on the morphology of the cells that look like signet rings under the microscope. This type of cancer is associated with a very poor outcome and seems to be more common in young people. However, very little is known about the molecular characteristics of SRCC and its development in different locations of the GI tract, essential pieces of knowledge that are needed to work towards better future treatment strategies.
Approach
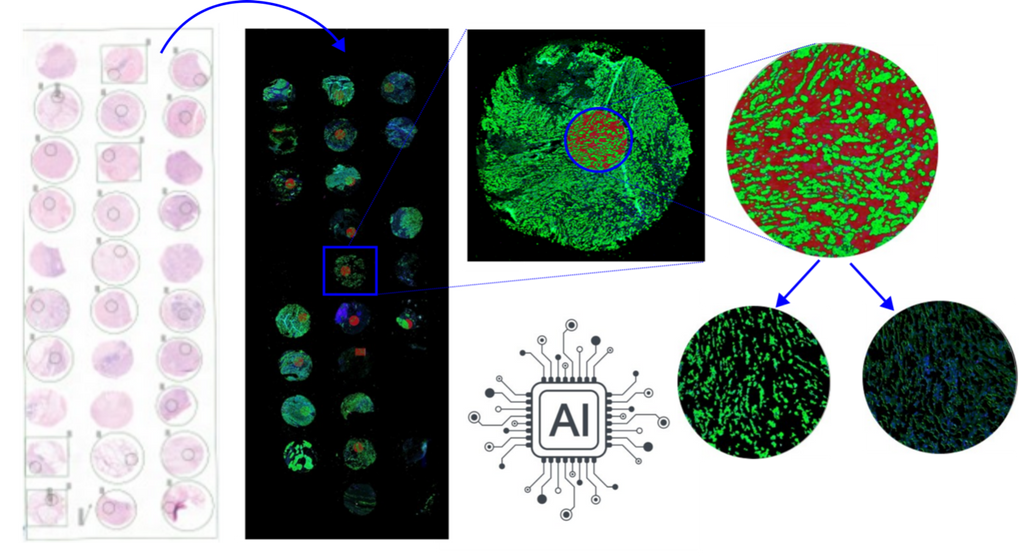
In this project, we will investigate the molecular profile of SRCC using histopathology data and spatial transcriptomics technology. We will develop image analysis and pattern recognition techniques using artificial intelligence models, to compare and contrast SRCC from the colon and stomach investigating their molecular components identifying patterns of gene expression.
Data
We will use 40 SRCC from the colon and 40 SRCC from the stomach, selected from the pathology archive of the Radboudumc. From the tumor tissue we will use: - Digital pathology whole-slide images (WSI) and tissue microarray (TMA) cores stained with hematoxylin and eosin (H&E) - TSO500 mutational analysis - NanoString GeoMx spatial transcriptomics of tumor and stroma
References
[1] Franko J, Le VH, Tee MC, Lin M, Sedinkin J, Raman S, Frankova D. Signet ring cell carcinoma of the gastrointestinal tract: National trends on treatment effects and prognostic outcomes. Cancer Treat Res Commun. 2021;29:100475. doi: 10.1016/j.ctarc.2021.100475. Epub 2021 Oct 11. PMID: 34655861.
Requirements
Students with a major in computer science, biomedical engineering, artificial intelligence, or a related area in the final stage of master level studies are invited to apply. Knowledge of the Python programming language is needed. Knowledge of R is a plus.
Information
- Project duration: 6 months
- Location: Radboud University Medical Center
- You will be part of the Diagnostic Image Analysis Group (DIAG) and Computational Pathology Group, whose research focus is the analysis of histopathological slides with deep learning techniques.
- You will have access to and work with a large GPU cluster.
- For more information, please contact Francesco Ciompi and Femke Doubrava-Simmer (femke.doubrava-simmer@radboudumc.nl)



