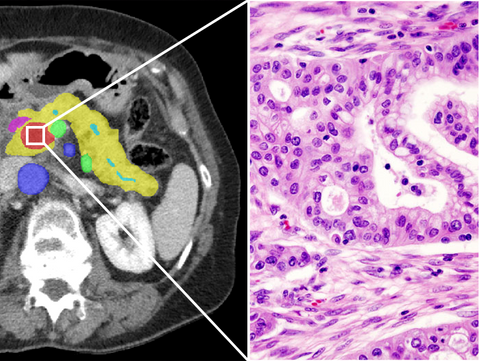
Clinical Problem
Pancreatic ductal adenocarcinoma (PDAC), usually called pancreatic cancer, is now the seventh leading cause of cancer-related deaths worldwide and will soon become the second leading cause of cancer-related death in Western society. With an average survival of 4.6 months, PDAC, which boasts a 5-year survival rate of less than 5% ,emerges as the deadliest cancer globally, resulting in patients to lose up to 98% of their healthy life expectancy.
Patient management for PDAC is mainly based on the well-known TNM staging system developed by the Union International Contre le Cancer (UICC), now in the 8th edition. This system stratifies patients by grading the tumor size (T), severity of the spread into regional lymph nodes (N) and the status of other distant metastatis (M), but despite being widely used, it is considered unreliable as patients with the same TNM stage often present different prognosis. Given the limitations of TNM staging for prognostication in clinical practice, there is a clear need for identifying reliable biomarkers that better correlate tumor characteristics with patient outcome.
Solutions
Recent advancements in using machine learning techniques in digitized pathology have paved the way for exploring quantitative morphological biomarkers in enhancing prognostic categorization for pancreatic cancer. In this endeavor, we will extract multiple AI-generated features and assess their correlation with patient outcomes to identify and rank dependable biomarkers. This undertaking entails the application and meticulous refinement of existing AI algorithms for pancreatic ductal adenocarcinoma (PDAC), specifically targeting parameters such as tumor-stroma ratio (TSR), mitotic count, and cell count. Additionally, we will automatically extract pertinent morphological characteristics, including tumor area and diameter, with each feature corresponding to a distinct quantitative measure. Following the initial phase, the extracted features will be subjected to correlation analysis in various experiments, such as predicting long-term versus short-term survival among patients.
Tasks
- Develop deep learning systems to quantify pathomics features in pancreatic cancer
- Apply existing deep learning models for the extraction of some biomarkers
- Use extracted features to build a prediction model of patient survival
- Deploy a public, open-source algorithm on Grand-Challenge.org
Requirements
- Students with a major in computer science, biomedical engineering, artificial intelligence, physics, or a related area in the final stage of master level studies are invited to apply.
- Affinity with programming in Python
- Interest in deep learning and medical image analysis
Information
- Project duration: 6 - 9 months
- Location: Radboud University Nijmegen Medical Center
- For more information, please contact Pierpaolo Vendittelli

